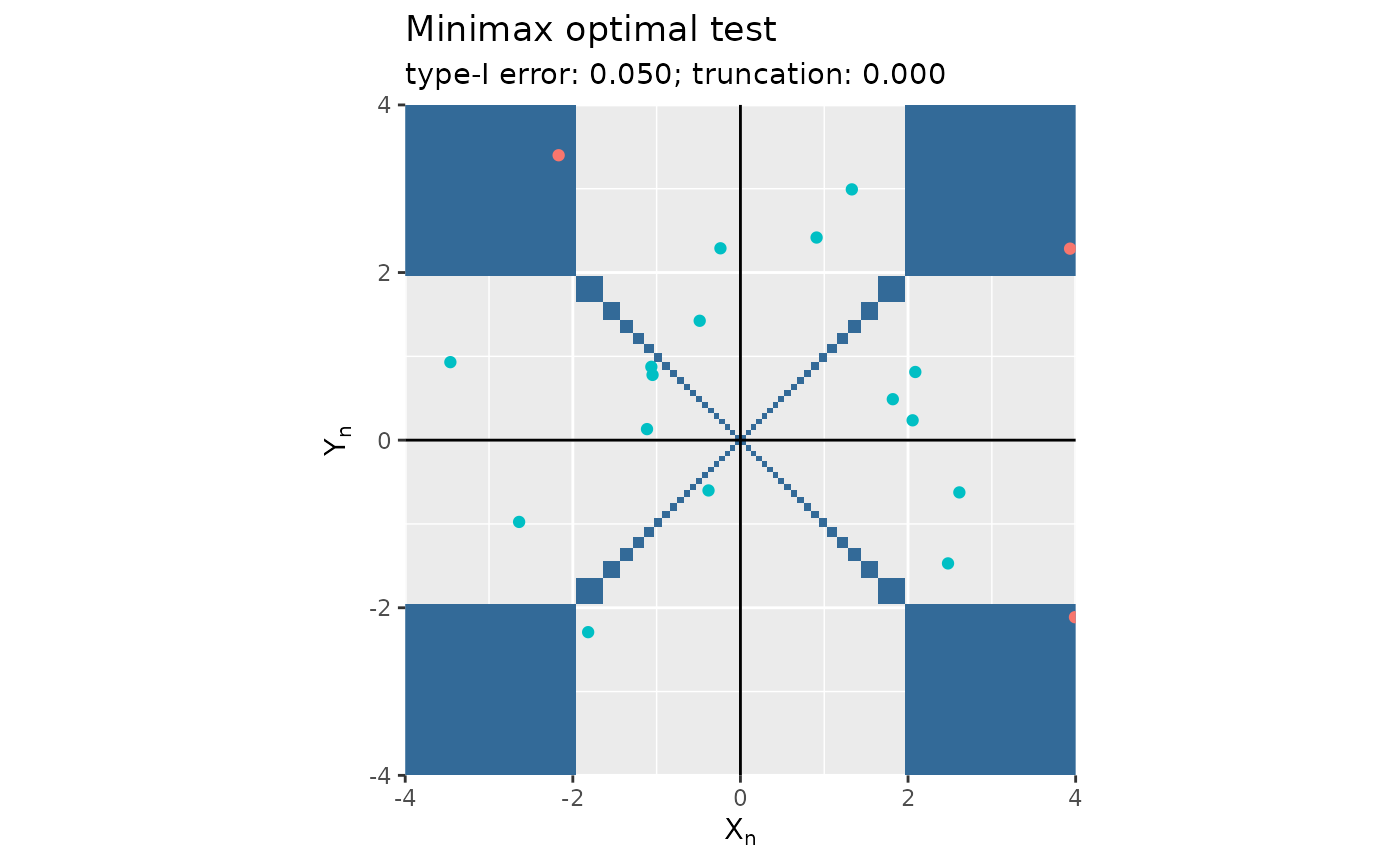
Minimax optimal testing
mediation_test_minimax.RdCarries out the minimax optimal test of the composite null "\(\delta_x \times \delta_y=0\)" against its alternative "\(\delta_x \times \delta_y\neq 0\)" based on the test statistic in the real plane.
Usage
mediation_test_minimax(
t,
alpha = 0.05,
truncation = 0,
sample_size = Inf,
compute_pvals = TRUE
)Arguments
- t
A
vectorconsisting of twonumerics, the test statistic in the real plane, or a 'n x 2'matrixof such test statistics.- alpha
A positive
numeric, the wished type-I error.- truncation
A nonnegative
numericused to bound the rejection region away from the null hypothesis space. Defaults to 0, in which case the rejection region is minimax optimal.- sample_size
An
integer(larger than one), the size of the sample used to derive the test statistic. Defaults to 'Inf', meaning that, under the null hypothesis, the test statistic is drawn from the \(N_2(0,I_2)\) law. If the integer is finite, then, under the null hypothesis, the test statistic is drawn from the product of two Student laws with 'sample_size-1' degrees of freedom.- compute_pvals
A
logicalindicating whether or not (conservative) p-values should be computed.
Value
A list, consisting of:
- t:
a
vectorof twonumerics, the test statistic, or a 'n x 2'matrixof such test statistics;- alpha:
a
numeric, the type-I error;- truncation:
a nonnegative
numeric, used to bound the rejection region away from the null hypothesis space- sample_size:
an
integer, the size of the sample used to derive the test statistic- decision:
a
vectoroflogicals,FALSEif the null hypothesis can be rejected for the alternative at level 'alpha' andTRUEotherwise;- pval:
a
vectorofnumerics, the (conservative) p-values of the tests;- method:
the
character"minimax".
Details
For details, we refer to the technical report "Optimal Tests of the Composite Null Hypothesis Arising in Mediation Analysis", by Miles & Chambaz (2024), https://arxiv.org/abs/2107.07575
See also
mediation_test_minimax_BH(), which builds upon the present function to implement a Benjamini-Hochberg procedure to control false discovery rate.
Examples
n <- 10
x <- MASS::mvrnorm(2 * n, mu = c(0, 0), Sigma = diag(c(1, 1)))
delta <- matrix(stats::runif(4 * n, min = -3, max = 3), ncol = 2)
epsilon <- stats::rbinom(n, size = 1, prob = 1/2)
delta <- delta * cbind(c(epsilon, rep(1, n)),
c(1 - epsilon, rep(1, n)))
x <- x + delta
(mt <- mediation_test_minimax(x, alpha = 1/20))
#> Testing the composite null 'delta_x * delta_y = 0' against its alternative 'delta_x * delta_y != 0':
#> * method:
#> minimax
#> * test statictic:
#> [,1] [,2]
#> [1,] -0.4338898 1.0794788
#> [2,] 0.3819511 -2.2583288
#> [3,] 1.5922643 -1.4707363
#> [4,] -1.0631020 0.3902375
#> [5,] -1.0487126 4.2130809
#> [6,] -2.1334481 0.2366963
#> ...
#> * wished type-I error:
#> [1] 0.05
#> * user-supplied truncation parameter:
#> [1] 0
#> * size of the sample used to derive the test statistic ('Inf' to use a Gaussian approximation; otherwise, use a product of Student laws):
#> [1] Inf
#> * decision [2 rejection(s) overall]:
#> cannot reject the null for its alternative with confidence 0.050
#> cannot reject the null for its alternative with confidence 0.050
#> can reject the null for its alternative with confidence 0.050
#> cannot reject the null for its alternative with confidence 0.050
#> cannot reject the null for its alternative with confidence 0.050
#> cannot reject the null for its alternative with confidence 0.050
#> ...
#> * (conservative) p-value:
#> [1] 0.66436847 0.70249763 0.09040863 0.69636097 0.29431040 0.81289242
#> ...
plot(mt)
#> Some points fall outside the range of the figure.